Biotechnology Team
Staff
- Team leader: Dr. Katarzyna Mikołajczyk, Ph.D.
- Dr. Laurencja Szała, Ph.D.
- Dr. Marcin Matuszczak, Ph.D.
- Dr. Agnieszka Łopatyńska, Ph.D.
- Dr. Joanna Wolko, Ph.D.
- Joanna Nowakowska, M.Sc.
- Katarzyna Kozłowska
- Julia Żok, M.Sc.
Research areas
- Mapping of winter oilseed rape chromosomes using DNA markers linked with quality traits.
- Development and application of functional genetic markers for marker assisted selection and marker assisted breeding (MAS and MAB) of the new oilseed rape breeding lines:
- A SCAR marker for the Rfo restorer gene applied in the ogura male-sterility system;
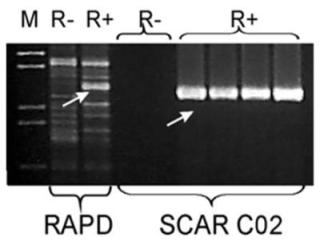 A SCAR marker
A SCAR marker
- A ‘multiplex PCR’ genetic assay for identification of breeding lines and recombinants with the Rfo restorer gene, as well as the ogura male-sterile cytoplasm in F1 hybrid breeding using ogura system;
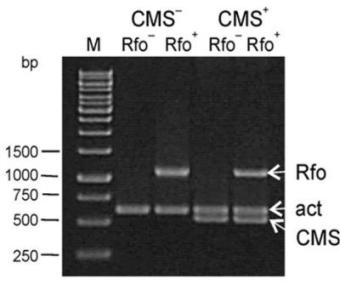 A ‘multiplex PCR’ genetic assay
A ‘multiplex PCR’ genetic assay
- Allele-specific functional SNaPshot markers for identification of non-mutated and mutant alleles of FAD3 desaturase genes in the A and C oilseed rape genomes, in low linolenic mutant lines and recombinants with double low lines;
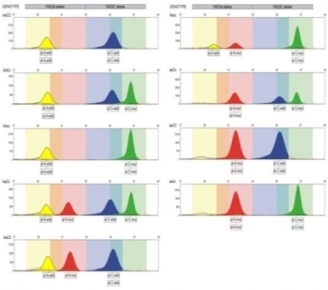 Allele-specific functional SNaPshot markers
Allele-specific functional SNaPshot markers
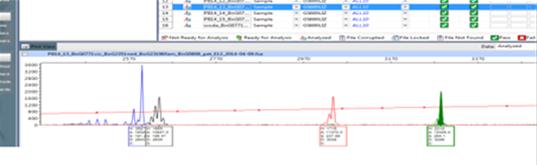
- A multiplex fluorescent PCR assay for monitoring Rfo restorer gene, the ogura male-sterile cytoplazm and also allelic forms of the FAD3 desaturase genes in the A and C oilseed rape genomes in breeding programs of new lines characterized by high yield and different composition of fatty acids in seed oil;
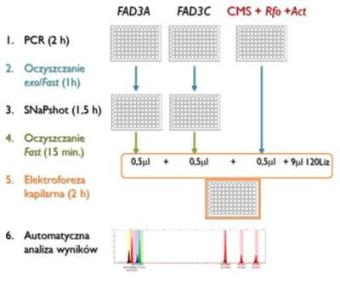 A multiplex fluorescent PCR assay
A multiplex fluorescent PCR assay
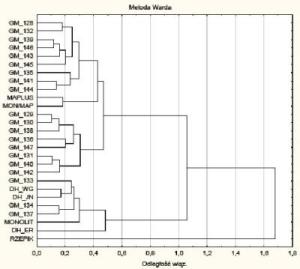
- Determination of microsatellite marker profiles for particular breeding forms of oilseed rape (fingerprinting) and assessment of genetic similarity among parental lines before crosses for broadening genetic diversity, as well as monitoring of genome rearrangements;
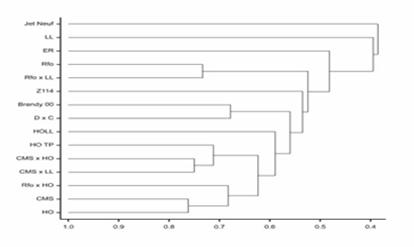 Identification of microsatellite markers
Identification of microsatellite markers
- Evaluation of genetic distance of parental lines using various forms of molecular markers for evaluation of hybrid performance in breeding programs.
- Application of molecular markers in hybrid varieties breeding of oilseed rape.
- Association studies (genome wide association studies, GWAS) using latest bioinformatics tools: next generation sequencing (Illumina), genotyping by sequencing methods in order to identify genes responsible for quality traits of oilseed rape.
- Application of winter oilseed rape (Brassica napus L.) doubled haploids (DHs) developed using isolated microspore culture for different breeding programs and genetic studies:
- Development of winter oilseed rape DH line mapping populations for association mapping and QTL genetic mapping;
- Production of DH lines with restorer gene for CMS ogura hybridization system;
- Phenotypic analysis of yield component traits in oilseed rape DH lines;
- Analysis of the content of seed compounds: fat, fatty acids, glucosinolates, bioactive compounds (tokochromanols, sterols, phenolic compounds) in DH lines;
- Statistical and genetic studies of quantitative traits in rapeseed as well as the analysis of the genotype × environment interaction.
 Production of doubled haploids of oilseed rape (Brassica napus L.)
Production of doubled haploids of oilseed rape (Brassica napus L.)
- Analysis of applied biotechnology methods to increase biodiversity in winter oilseed rape:
- Transformation of haploid cells using direct co-cultivation of androgenic embryos with Agrobacterium tumefaciens;
- Production of doubled haploids derived from transformed microspores and androgenic embryos;
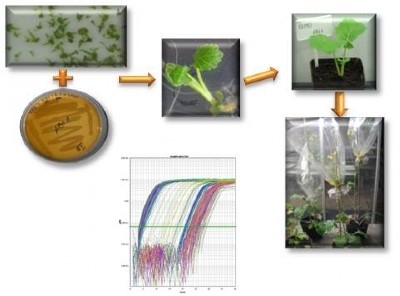 Genetic transformation of microspore-derived embryos
Genetic transformation of microspore-derived embryos
- Resynthesis of Brassica napus through interspecific crosses of selected subspecies of Brassica rapa and Brassica oleracea;
- Cytogenetic and molecular analysis of resynthesized oilseed rape embryos;
- Development of semi-RS DH lines with double low quality of winter oilseed rape.
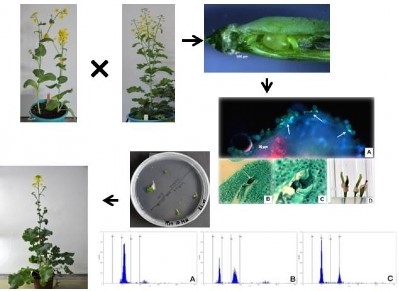 Resynthesis of Brassica napus L.
Resynthesis of Brassica napus L.
- White mustard (Sinapis alba L.) androgenesis in the culture of isolated microspores.
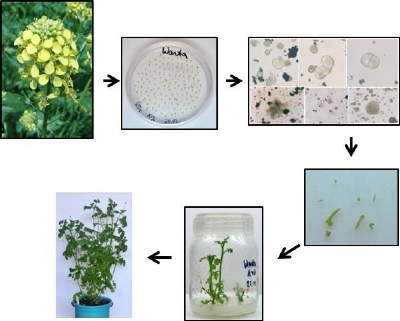 In vitro androgenesis of Sinapis alba L.
In vitro androgenesis of Sinapis alba L.